The exact order of nucleotides (guanine (G), adenine (A), thymine (T), and cytosine (C)) in DNA amplified by PCR from viral genomic material is determined by DNA sequencing. Although presence of the virus can be confirmed by PCR, sequence data is useful for identifying viral gene relatedness and variation, and for understanding viral evolution. This information can be used to examine epidemiological relationships and to detect the emergence of new viral species.
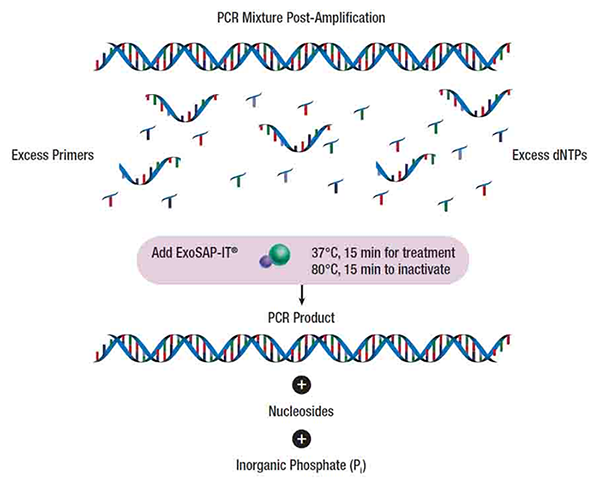
- Exonuclease I to remove residual single-stranded primers and any extraneous single-stranded DNA produced in the PCR.
- (recombinant) Shrimp Alkaline Phosphatase to remove the remaining dNTPs from the PCR mixture. The reaction proceeds for 15 minutes at 37 deg C. A termination reaction (heating at 80 deg C) takes a further 15 minutes. The sample is now ready to be sequenced.
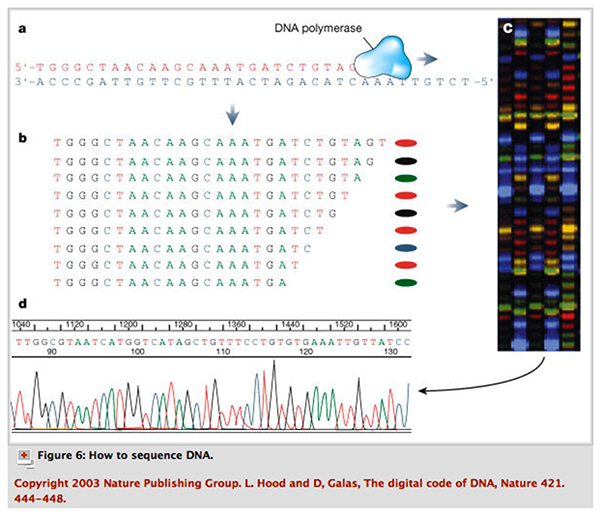
Cycle sequencing using fluorescence-labeled dideoxynucleotides is a commonly used method to sequence DNA. It is based on the Sanger sequencing process. Cycle sequencing uses a thermostable DNA polymerase, which allows the reaction to be repeated many times in the same tube. The reaction cycles between :
- Denaturation of the target DNA at 94 deg C.
- Primer annealing to the denatured or separated DNA strands at 50 deg C.
- Polymerization by the DNA polymerase at 72 deg C.
The reaction occurs in a thermocycler (see previous module on PCR). The reaction mix includes:
- DNA template.
- Primer.
- Thermostable DNA polymerase.
- Unlabeled dNTPs.
- Fluorescently labeled ddNTPs. ddNTPs lack a 3’OH group, and nothing can be added to the growing DNA chain once a ddNTP has been added: the sequence is terminated.
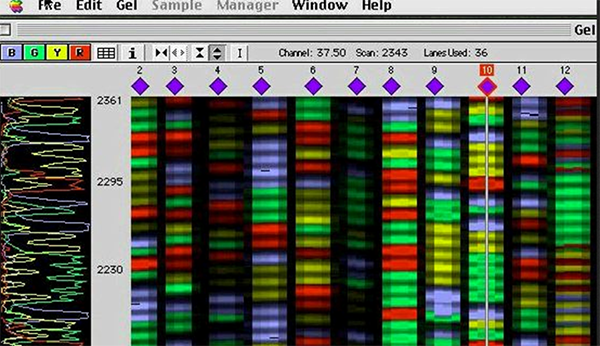
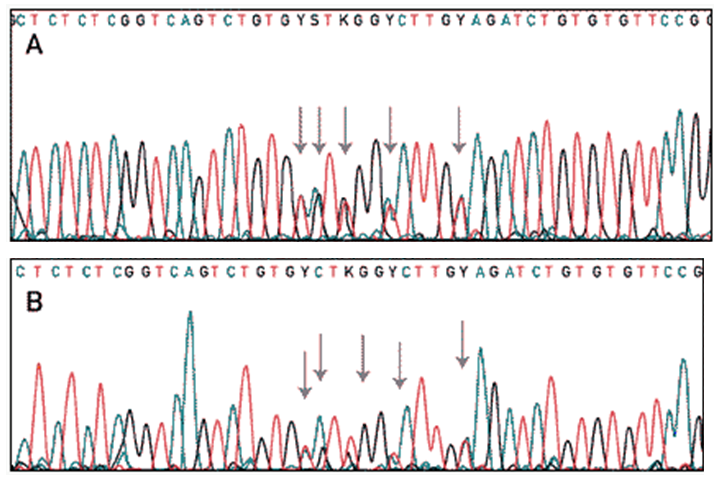
When the reaction is complete, the product which contains many DNA fragments of different sizes, is loaded onto a polyacrylamide gel in a sequencing instrument. The sequencer will detect the fluorescent dye as the DNA fragments migrate through the gel towards the anode, and the computer complies the data into an image of a gel. The colours are then read as bases and a chromatogram of the piece of DNA being sequenced is produced. Each of the four colours represents a different base, and by translating the colours to bases, the DNA sequence can be determined. The computer only translates or ‘calls’ strong signals. When there are conflicting signals the ambiguous base is designated ‘Y’. Here is a summary of cycle sequencing